Research Summary: Sequencing Lifeforms in Lake Vostok Ice Cores
1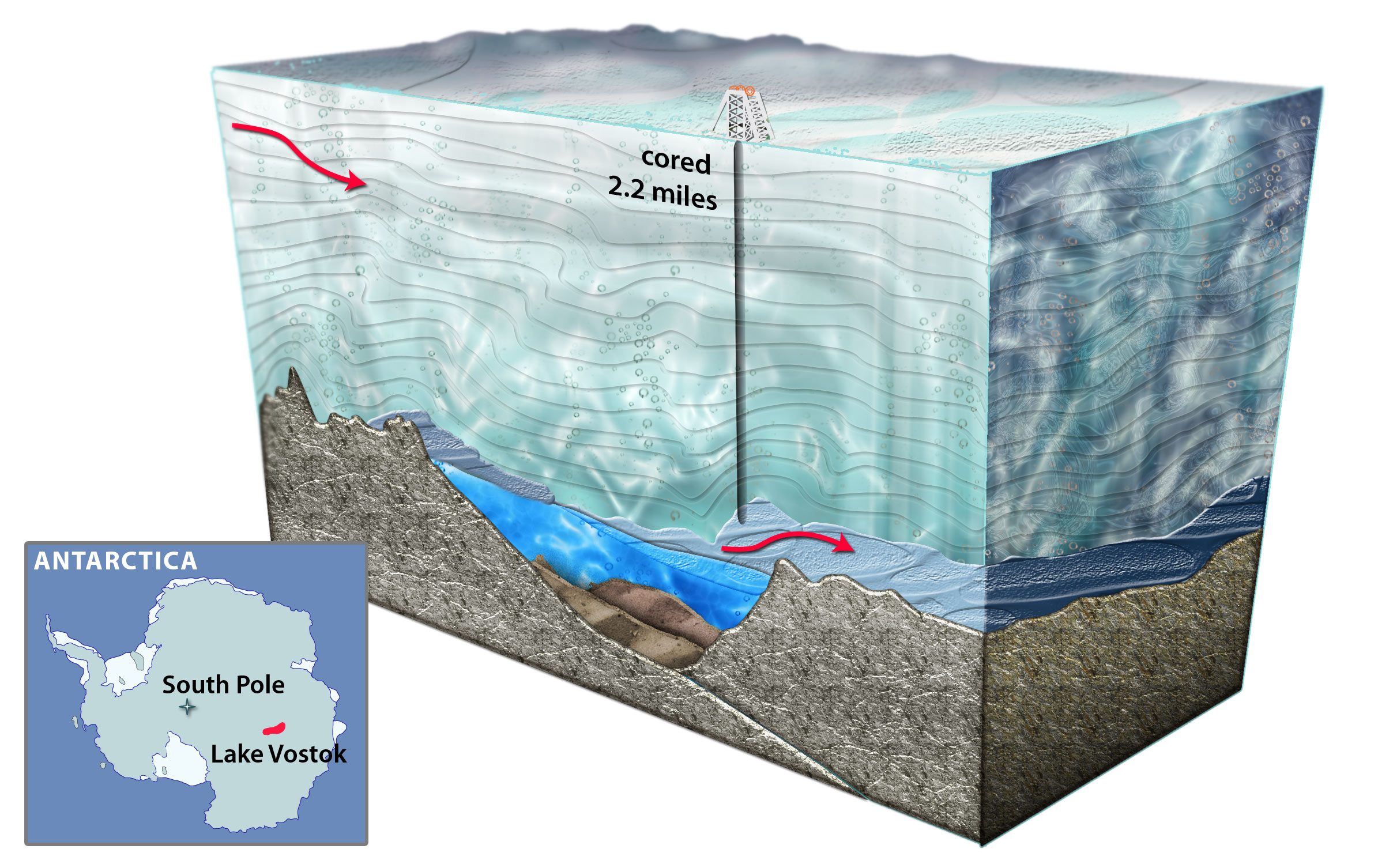
Three dimensional model of Lake Vostok drilling. (Credit: National Science Foundation)
Lake Vostok is the largest of nearly 400 subglacial lakes that have been found in Antarctica, at least some of which are connected by subglacial rivers and streams. As the overriding 4 km-thick glacier moves across Lake Vostok at a rate of 3 m yr−1, lake water freezes (i.e., accretes) to the bottom of the glacier creating a lake-water record that is a linear and temporal representation of the surface contents of the lake. At the Vostok 5G ice core drill site on the far side of the lake (relative to the glacier entry site), the meteoric ice is 3538 m thick, and the accretion ice is more than 231 m thick. At that location, the glacier has passed over an embayment, near a peninsula/ridge, and then over part of the southern main lake basin. Ice that accreted in the vicinity of the embayment has relatively high concentrations of ions, organic carbon, biomass and mineral inclusions (termed “type I” – or type 1 – accretion ice). Over the majority of the lake, the accretion ice contains far fewer mineral inclusions, as well as lower concentrations of ions, organic carbon, and biomass. This relatively clear ice is known as “type II” (or type 2) accretion ice.
Several studies have described the biotic and abiotic components from Lake Vostok accretion ice core sections. Mean cell concentrations in the accretion ice ranged from <1 to several hundred cells ml−1. In most studies, the highest concentrations of cells and highest sequence diversities were reported in accretion ice from within or near the embayment in the southwest corner of the lake near the entry point of the glacier. Additionally, we and others demonstrated that the concentrations of total cells and viable cells were higher in the accretion ice than in the meteoric ice above. In the same studies, we cultivated, sequenced (rDNA regions) and identified 18 unique isolates of Bacteria and 31 unique isolates of Fungi from the same Lake Vostok accretion ice core sections. All were phylogenetically closest to species from aquatic, lake/ocean sediment, cold, polar and/or deep-sea environments. The compilation of results suggests that organisms are living and reproducing in Lake Vostok. For the present metagenomic/metatranscriptomic study, we examined two ice core sections that accreted in the vicinity of the embayment (at depths 3563 and 3585 m), and two ice core sections that accreted over the southern main lake basin (at 3606 and 3621 m). The results of this research provide a detailed view of the potential life in Lake Vostok.
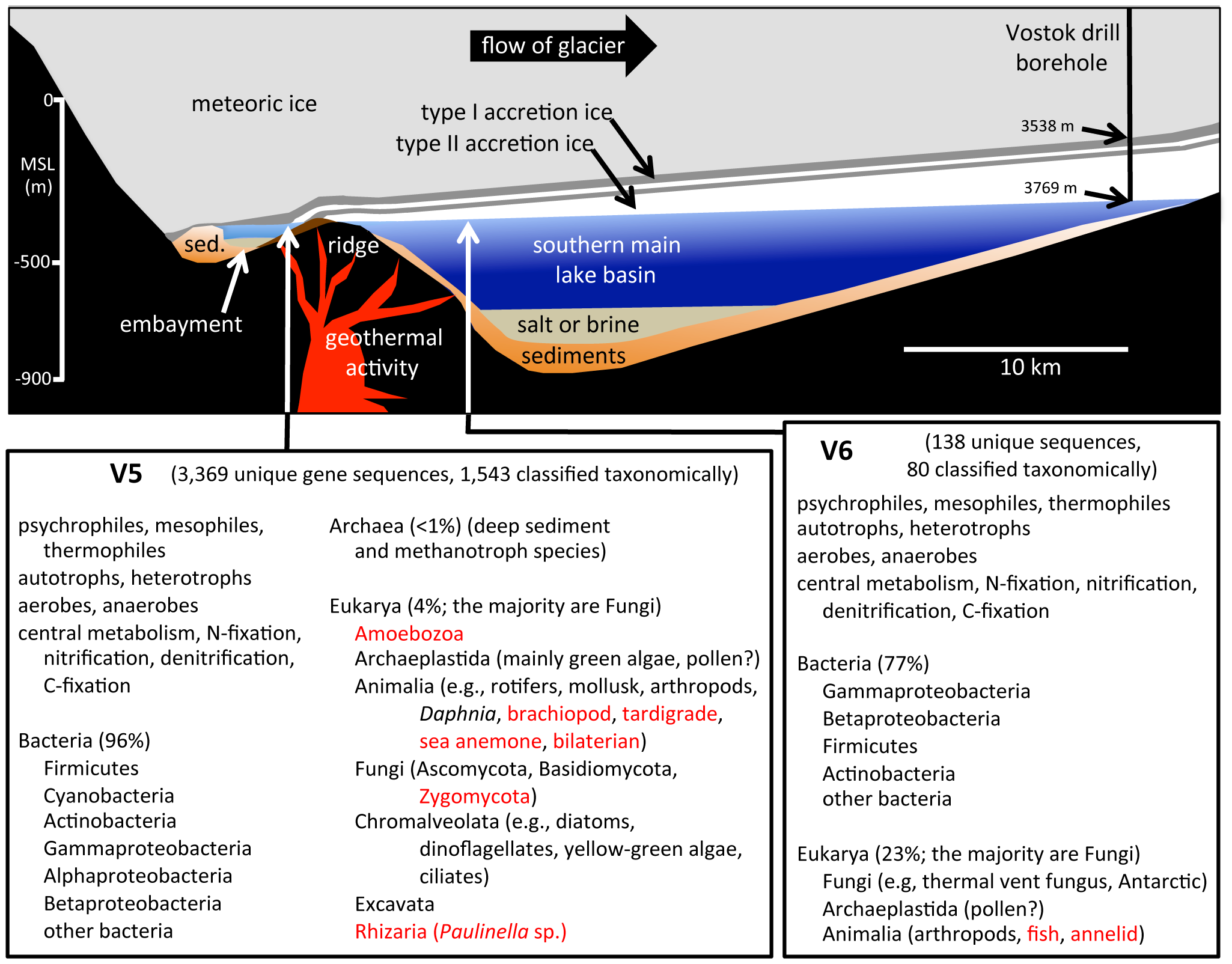
Schematic cross-section of Lake Vostok drawn to scale (based on a radar study of Lake Vostok along the glacial flow line to the ice core drill site). Summary of V5 and V6 samples below.
Methods
The ice core sections were selected from the USGS NICL (United States Geological Survey, National Ice Core Laboratory, Denver, CO), and were shipped frozen to our laboratory. Sample V5 consisted of Vostok 5G core sections at 3563 and 3585 m (type I ice), corresponding to ice that accreted in the vicinity of the embayment. Sample V6 included core sections 3606 and 3621 m (type II ice), corresponding to ice that accreted over a portion of the southern main basin of Lake Vostok. Each core section was surface sterilized using a method that had been previously developed, tested, described and utilized. Briefly, quartered ice core sections, 6–16 cm in length (total volume approximately 125 ml), were immersed in a 5.25% sodium hypochlorite solution (pre-chilled to 4°C for at least 2 h) for 10 s followed by three rinses with 800 ml of sterile water (4°C, 18.2 MΩ, <1 ppb total organic carbon, autoclaved). The sections were then melted in sterile funnels and meltwater was collected in 50 ml aliquots. Each aliquot comprised a “shell” of meltwater corresponding to the outer portion, and sequentially more interior portions of the ice core section. The meltwater was then frozen at −20°C. A total of 250 ml of meltwater was used for each sample (125 ml from each ice core section). The meltwater samples were filtered sequentially through 1.2, 0.45 and 0.22 µm Durapore filters (Millipore, Billerica, MA). The filtered meltwater was subjected to ultracentrifugation at 100,000×g (aseptically) for 16 hours to pellet cells and nucleic acids. The filtered meltwater contained small cells, cell debris, viruses and biomolecules (including RNA and DNA) from single-celled and multicellular organisms. Two control samples (purified water, 18.2 MΩ, <1 ppb total organic carbon; and the same water, autoclaved and subjected to concentration by ultracentrifugation) also were processed using the same protocols. The V5, V6, and control samples were ultracentrifuged on different days to lessen potential cross-contamination. Pellets were rehydrated in 50 µl of sterile 0.1X TE (1 mM Tris [pH 7.5], 0.1 mM EDTA). Nucleic acid extraction was performed using MinElute Virus Spin Kits (QIAGEN, Valencia, CA) according to the manufacturer’s instructions and eluted in 150 µl AVE buffer (with 0.04% sodium azide). The eluted nucleic acids were allowed to precipitate overnight at −20°C with 0.5 M NaCl in 80% ethanol. They were then pelleted by centrifugation at 16,000×g for 15 min, washed with cold 80% ethanol and centrifuged at 16,000×g for 5 min. They were dried under vacuum, and were resuspended in 15 µl 0.1X TE. All glassware, tubes, and piptet tips were treated with RNase Away (Life Technologies, Grand Island, NY) and autoclaved prior to use. Solutions and reagents were autoclaved (except those purchased as reaction mixes).
Additional methods: cDNA synthesis; adapter ligation and chromatography; addition of 454 A and B sequences by PCR amplification; and genome sequence analysis.
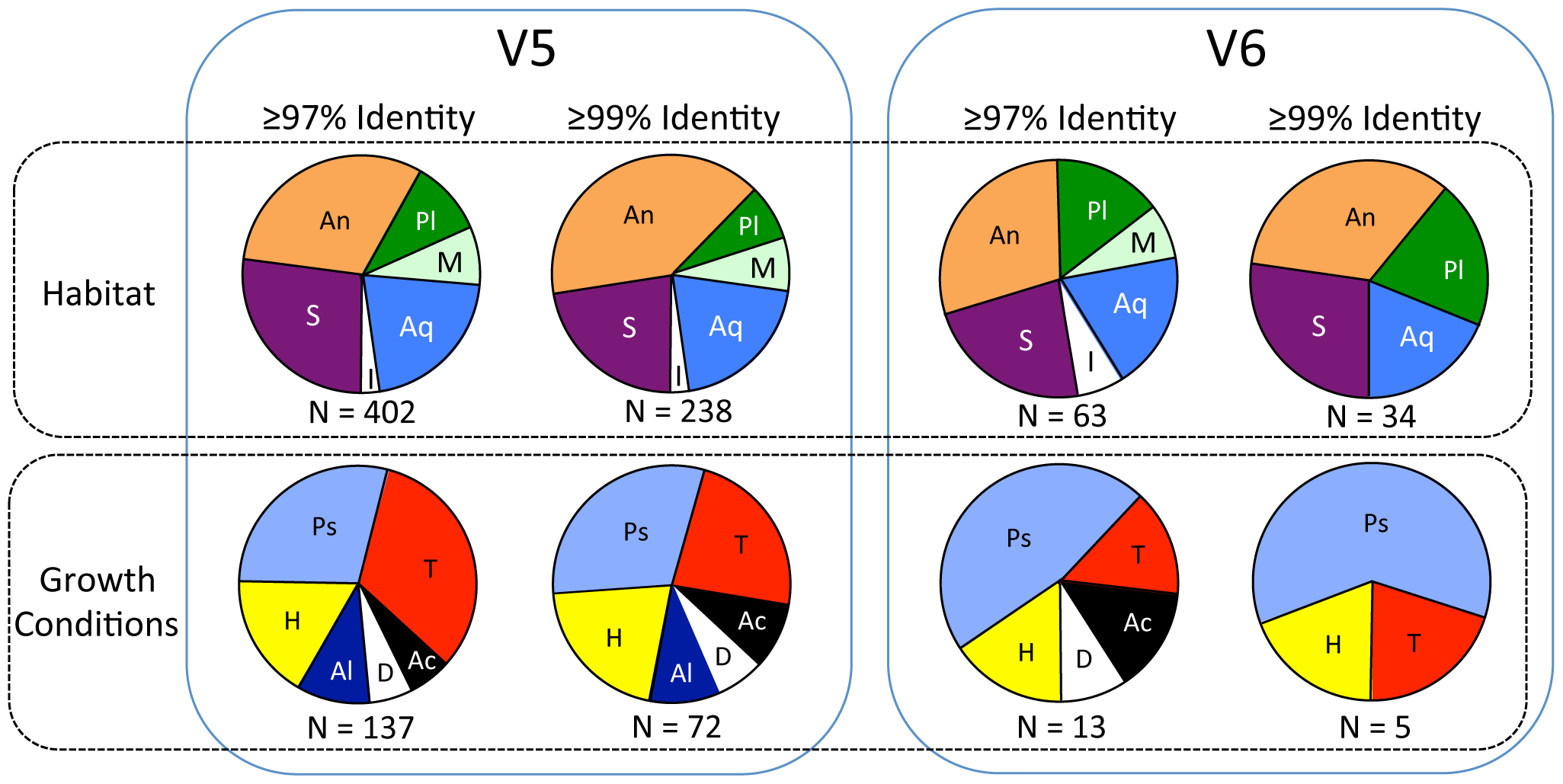
Summary of proportions of sequences in V5 (left) and V6 (right) categorized by habitat (upper row) and growth conditions (lower row) based on species with highest sequence identities. Habitat abbreviations: An = animal associated (most are also found in soils and/or water); Aq = aquatic; I = ice, glaciers and/or polar; M = marine; Pl = plant associated (most are also found in soils and water); S = soils or sediments. Growth conditions abbreviations: Ac = acidophilic or acid tolerant; Al = alkaliphilic or alkali tolerant; D = desiccation resistant; H = halophilic or halotolerant; Ps = psychrophilic or psychrotolerant; T = thermophilic or thermotolerant. Number of sequences (N) used for each is indicated below each pie chart.
Results
A total of 36,754,464 bp of sequence data was obtained from sample V5 that included 94,728 high quality 454 sequence reads, with a mean length of 388 bp. For the V6 sample, a total of 1,170,900 bp of sequence data was obtained that included 5,204 high quality reads, with a mean length of 225 bp. The lower quantity of sequence data for V6 might have resulted from lower nucleic acid concentrations (as we repored previously), as well as higher degrees of degradation. Overall, approximately 15% of the sequences were unique, while the remaining 85% were additional copies from the unique set of sequences. A total of 3,369 unique sequences were derived from V5, of which 1,543 could be taxonomically classified, and 138 unique assembled sequences were derived from V6, of which 80 could be taxonomically classified. Approximately 94% of the unique sequences in V5 and 83% in V6 were from Bacteria. Only two unique Archaea sequences were found (both in V5), and they were most similar to methanotrophs from cold deep-ocean sediments. The remainder were Eukarya (4% in V5 and 17% in V6), including more than 150 unique sequences from multicellular organisms, most of which were Fungi. In general, the taxa were similar to organisms specific to lakes, brackish water, marine environments, soil, lake sediments, deep-sea sediments, deep-sea thermal vents, animals and plants. Sequences from autotrophs and heterotrophs were present.
Acknowledgements
We thank the USGS National Ice Core Laboratory (NICL), NICL-Science Management Office (NICL-SMO) and the National Science Foundation for allowing us access to the ice cores used in this study. We thank Dr. Li-Jun Ma for help in the initial stages of this study. We thank the staff at the Ohio Super Computer for allowing us the use of their computational resources.
Full study and references published July 2013 in PLOS ONE (Online, Open Access). Additional study authors include Yury Shtarkman, Zeynep Kocer, Robyn Edgar, Ram Veerapaneni, Tom D’Elia and Paul Morris.
Selected References
- Kapista A, Ridley JF, Robin GDeQ, Siegert MJ, Zotikov I (1996) Large deep freshwater lake beneath the ice of central Antarctica. Nature 381: 684–686.
- MacGregor JA, Matsuoka K, Studinger M (2009) Radar detection of accreted ice over Lake Vostok, Antarctica. Earth Planet Sci Lett 282: 222–233.
- Masalov VN, Lukin VV, Shermetiev AN, Popov SV (2001) Geophysical investigation of the subglacial Lake Vostok in Eastern Antarctica. Doklady Earth Sci 379A: 734–738.
- Studinger M, Karner GD, Bell RE, Levin V, Raymond CA, et al. (2003) Geophysical models for the tectonic framework of the Lake Vostok region East Antarctica. Earth Planet Sci Lett 216: 663–677.
- Wright A, Siegert MJ (2011) The identification and physiographical setting of Antarctic subglacial lakes: An update based on recent discoveries. Geophysical Monograph Ser 192: 9–26.
- Wingham D, Siegert M, Shepherd A, Muir AS (2006) Rapid discharge connects Antarctic subglacial lakes. Nature 440: 1033–1036.
- Jouzel J, Petit JR, Souchez R, Barkov NI, Lipenkov VY, et al. (1999) More than 200 meters of lake ice above subglacial Lake Vostok, Antarctica. Science 286: 2138–2141.
- Bell R, Studinger M, Tikku A, Castello JD (2005) Comparative biological analyses of accretion ice from subglacial Lake Vostok. In: Castello JD, Rogers SO, editors. Life in Ancient Ice. Princeton, NJ: Princeton University Press. 251–267.
- Gramling C (2012) A tiny window opens into Lake Vostok, while a vast continent awaits. Science 335: 788–789.
- Siegert MJ, Ellis-Evans JC, Tranter M, Mayer C, Petit J, et al. (2001) Physical, chemical and biological processes in Lake Vostok and other Antarctic subglacial lakes. Nature 414: 603–609.














[…] Lake Vostok’s lifeforms are any indication, this new lake may contain a large number of bacteria similar to those found in […]